Members of the ADDovenom consortium at University of Liège and Liverpool School of Tropical Medicine have co-authored an article, published by Toxins on 25 May 2023, which is based on their collaborative work on the project.
Next-Generation Sequencing for Venomics: Application of Multi-Enzymatic Limited Digestion for Inventorying the Snake Venom Arsenal was written by Fernanda Gobbi Amorim, Damien Redureau, Thomas Crasset, Lou Freuville, Dominique Baiwir, Gabriel Mazzucchelli, Stefanie K. Menzies, Nicholas R. Casewell and Loïc Quinton.
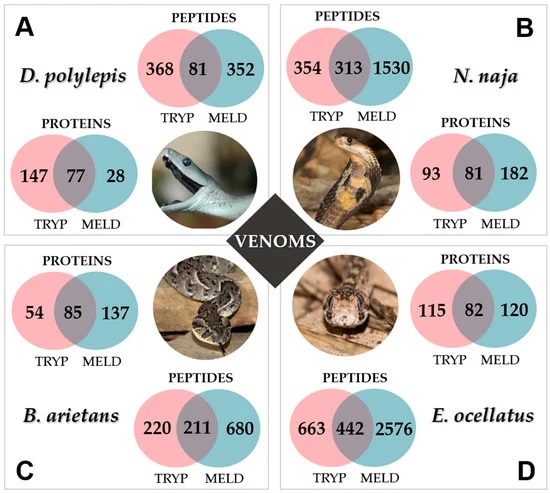
Their research examined the “application of a new generation of proteomic methodology to deeply characterize complex protein mixtures. The new approach, combining a synergic multi-enzymatic and a time-limited digestion (MELD), is a versatile and straightforward protocol previously developed by our group. The higher number of overlapping peptides generated during MELD increases the quality of downstream peptide sequencing and of protein identification. In this context, this work aims at applying the MELD strategy to a venomics purpose for the first time, and especially for the characterization of snake venoms.”
The results proved that MELD was successful in improving “the number of sequenced (de novo) peptides and identified peptides from protein databases, leading to the unambiguous identification of a greater number of toxins and proteins. For each venom, MELD was successful, not only in terms of the identification of the major toxins (increasing of sequence coverage), but also concerning the less abundant cellular components (identification of new groups of proteins). In light of these results, MELD represents a credible methodology to be applied as the next generation of proteomics approaches dedicated to venomic analysis. It may open new perspectives for the sequencing and inventorying of the venom arsenal and should expand global knowledge about venom composition.”
(Fernanda gave an online presentation on ‘Next-Generation Sequencing for Venomics: Application of Multi-Enzymatic Limited Digestion for Inventorying the Snake Venom Arsenal’ at the LPMHealthcare Venoms and Toxins Conference in August 2021).
Next-Generation Sequencing for Venomics: Application of Multi-Enzymatic Limited Digestion for Inventorying the Snake Venom Arsenal
Fernanda Gobbi Amorim, Damien Redureau, Thomas Crasset, Lou Freuville, Dominique Baiwir, Gabriel Mazzucchelli, Stefanie K. Menzies, Nicholas R. Casewell and Loïc Quinton
Toxins 2023, 15(6), 357; https://doi.org/10.3390/toxins15060357